Processing...
Choose a Disease:
Processing...
Description of the disease
Processing...
What is a SnP site?
A single nucleotide polymorphism (SnP) is the most common type of genetic variation between people. They occur on average once in every 1000 nucleotides (Roughly 4 million SnPs in total). As an example, a SNP may replace the nucleotide cytosine (C) with the nucleotide thymine (T) in a certain stretch of DNA. They can act as biological markers which help locate genes that are related to diseases. When SnPs occur with in or near a gene they may play a role in the disease by affecting the genes function.
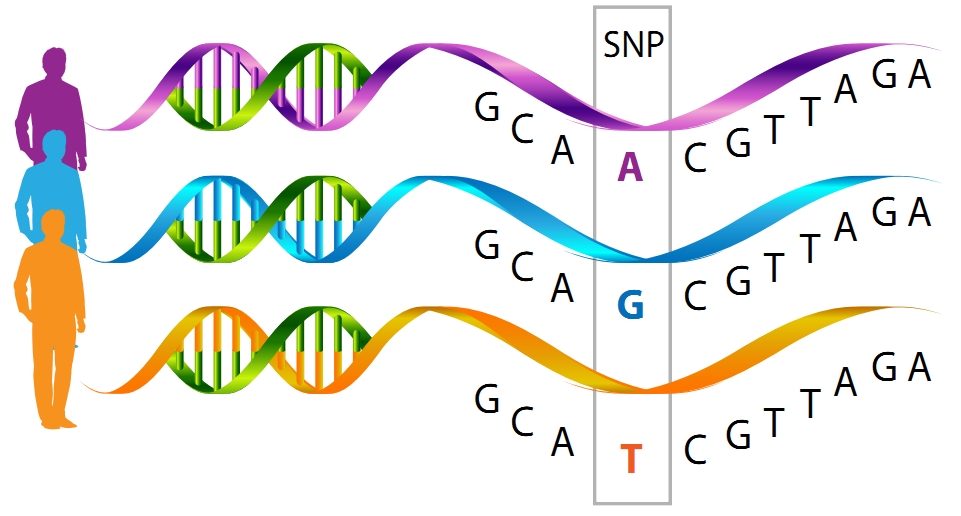
Most SNPs have no effect on health or development. Some of these genetic differences, however, have proven to be very important in the study of human health. Researchers have found SNPs that may help predict an individual’s response to certain drugs, susceptibility to environmental factors such as toxins, and risk of developing particular diseases. SNPs can also be used to track the inheritance of disease genes within families. Future studies will work to identify SNPs associated with complex diseases such as heart disease, diabetes, and cancer. For example, a single-base mutation in the APOE (apolipoprotein E) gene is associated with a lower risk for Alzheimer's disease.
A. Wolf, R. Caselli, E. Reiman and J. Valla, Neurobiology of Aging, 2013, 34, 1007-1017.
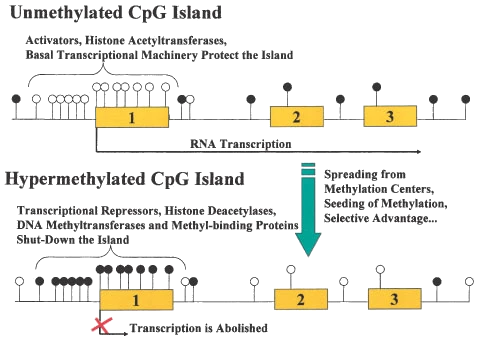
What is a CpG Island?
CpG Islands are important because they represent areas of the genome that have for some reason been protected from the mutating properties of methylation through evolutionary time (which tend to change the G in CpG pairs to an A). Often, they point to the presence of an important piece of intergenic DNA, such as that found in the promoter regions of genes where transcription factors bind. . In cancers, loss of expression of genes occurs about 10 times more frequently by hypermethylation of promoter CpG islands than by mutations.
How does it work?
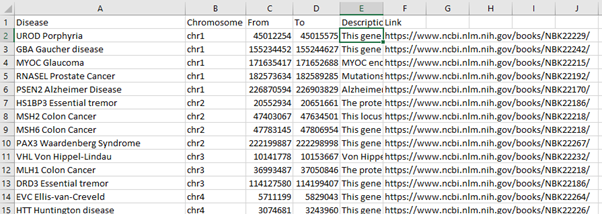
Once a disease has been selected and the run button is pressed this is connected to a spreadsheet which contains information about where to find this disease within our genes. The information on the spreadsheet was collected from the National Center for Biotechnology.
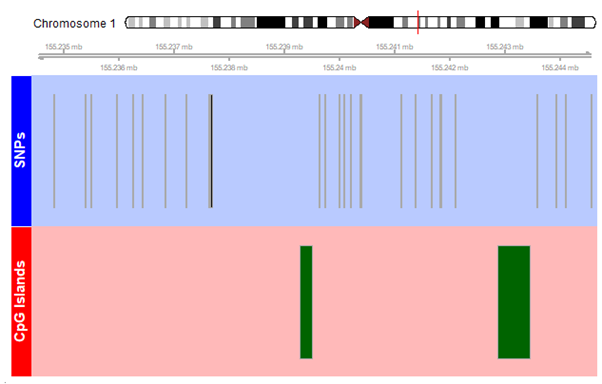
From this location it is then sent to the UCSC genome browser where it collects the SnP and CpG data for that location before the programme visualises the SnP and CpG island data using GViz a Bioconductor package for R.
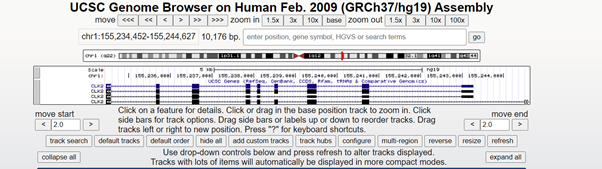
The lines on the SnP track show sites that have a minor allele frequency of at least 1% and are mapped to a single location in the reference genome assembly The highlighted areas on the CpG island are where there is a GC content of higher than 50% for greater than 200bp. The red line on the chromosome highlights the area in which the disease is located, and what you are viewing on the tracks is data within the highlighted region.